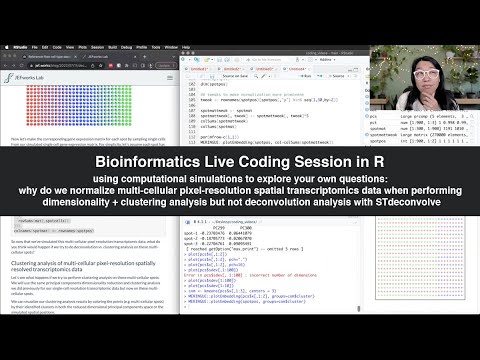
Live R Coding Session - single cell spatial transcriptomics data visualization in base R

Live R Coding Session - Spatial transcriptomics data analysis with STdeconvolve and SpotCleanПодробнее

10x Visium spatial transcriptomics data analysis with STdeconvolve in RПодробнее

Label single-cell RNA data automatically with SingleR in RПодробнее

Spatial Transcriptomics Intro - Part 1: What Is Spatial TranscriptomicsПодробнее

SpatialExperiment infrastructure for spatially resolved transcriptomics data in R usiПодробнее

Spatially-resolved transcriptomics analysis with R/Bioconductor and beyondПодробнее

Introduction to single-cell RNA-Seq and Seurat | Bioinformatics for beginnersПодробнее
Stereopy as an Advanced Tool for Interpreting Spatial Transcriptomics DataПодробнее

W31: Spatial Transcriptomics – Day 1Подробнее
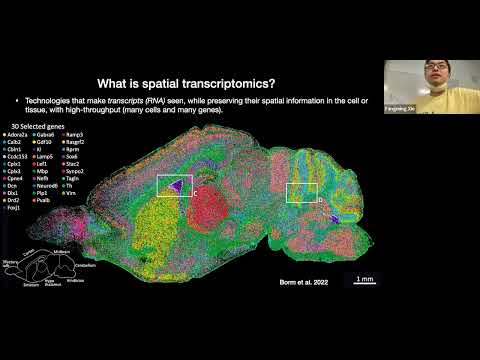
Live R Coding Session - normalizing spatial transcriptomics data for clustering vs deconvolutionПодробнее